Neutroner sonderer biologiske materialer for at få indsigt i COVID-19-virusinfektion
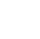
Den nye coronavirus-membran (lyseblå) og den humane cellemembran (orange) smelter sammen, når den virale S2-underenheds fusionspeptid (lilla pile) indsættes i cellemembranen, og en anden komponent af S2-underenheden (lilla og grøn) folder sig til dannelse en stram struktur, som vist øverst til højre. I modsætning, som illustreret nederst til højre, fusionshæmmere er designet til at forhindre virusinfektion ved at forstyrre denne proces. Kredit:ORNL/Jill Hemman
SARS-CoV-2, coronavirus, der er ansvarlig for sygdommen COVID-19, inficerer verden i høj fart. At forstå, hvordan denne infektion virker på molekylært niveau, kan hjælpe eksperter med at finde måder at moderere eller stoppe spredningen.
Et team af forskere ved Department of Energy's (DOE's) Oak Ridge National Laboratory (ORNL) bruger neutronreflektometri til at gøre netop det.
Neutroner er i stand til at sondere biologiske materialer under fysiologiske forhold uden at beskadige dem. Ved at udnytte disse egenskaber, forskerne kan måle virusets infektionsdynamik, når det sker.
Deres mission er at få et detaljeret kig på nogle af de første stadier af infektion, der opstår ved cellemembranen. Disse resultater vil hjælpe holdet med at teste antivirale lægemiddelkandidater, der kan forstyrre denne proces. Data opnået fra disse eksperimenter kunne også informere andre undersøgelser fokuseret på at udvikle terapeutiske midler og vacciner.
Forskerne fokuserer deres analyse på SARS-CoV-2 spike proteiner, modhagelignende strukturelle proteiner, der dækker overfladen af virussen og udløser infektionsprocessen. Spikeproteinet binder sig til en receptor på værtscellens ydre lag og letter fusion mellem virus- og cellemembraner, tillader virus at komme ind i cellen og frigive dets genetiske materiale. Cellens proteinfremstillingsmaskineri bruger derefter denne genetiske information til at lave nye kopier af virussen.
Når SARS-CoV-2 kaprer en værtscelle, dets spidsprotein deler sig i to underenheder, kaldet S1 og S2. De to dele er begge afgørende for infektion. S1-underenheden indeholder et receptorbindingsdomæne, der genkender og låser på en cellereceptor. Cellereceptorer er proteiner indlejret i cellemembranen, som kan binde til specifikke molekyler uden for cellen. Denne forbindelse kan få komponenterne til at ændre form, hvilket igen kunne inducere kaskadeændringer i cellen. For SARS-CoV-2-spidsproteinet, denne forbindelse aktiverer S2-underenheden, som hjælper virussen med at flette sin membran med cellens. Derfor, spidsproteinets funktion ligner at åbne en låst dør, med S1 som nøglen der låser døren op og S2 som kraften der skubber døren op.
Lær af tidligere epidemier
Den overordnede struktur af SARS-CoV-2 spike-proteinet ligner meget SARS-CoV, en tidligere coronavirus, der forårsagede alvorligt akut respiratorisk syndrom (SARS), og denne lighed hjalp teamet med at udvikle deres forskningsstrategi.
S1-underenheden er i fokus for mange lægemiddeludviklingsstudier, da denne del af spidsproteinet har vist sig at fremkalde et immunrespons i menneskekroppen. Imidlertid, tidligere SARS-CoV-undersøgelser viste, at S1-underenheden oplever høje mutationsrater, tillader virussen at unddrage sig antistof-baserede behandlinger, samtidig med at dens evne til at inficere celler bevares. "Dette er den lektie, vi har lært af den oprindelige SARS-epidemi, " sagde Minh Phan, en postdoktoral forskningsassistent ved ORNL og hovedefterforsker af dette projekt.
Phan og hans kolleger studerer S2-underenheden, fordi denne komponent af spikeproteinet ikke muterer så hurtigt. Behandlinger, der viser sig at være vellykkede med at hæmme S2-funktionen, kan forblive effektive i længere tid.
En nanoskala visning af coronavirus
For bedre at forstå dynamikken i virale S2-underenheder og værtscellemembraner, forskerne anvender væskereflektometeret (LIQREF) ved ORNL's Spallation Neutron Source (SNS). Ved at måle, hvordan neutroner reflekterer i forskellige vinkler, når de passerer gennem forskellige typer stof, instrumentet kan hjælpe med at belyse strukturen af biologiske materialer i molekylær skala.
Holdet syntetiserede først en lipidmembran, der efterligner den ydre membran af celler, der beklæder overfladerne inde i menneskets lunger, hvor denne virusinfektion kan finde sted. De identificerede, hvordan lipiderne var organiseret i membranen, og hvordan dette arrangement ændres, når membranerne udsættes for forskellige forhold, såsom temperatur, tryk, og surhedsgrad.
Ved LIQREF instrumentet, forskerne spredte lipidmembranen oven på et tyndt lag vand i et apparat kaldet et Langmuir trug. De introducerer derefter S2-underenheden til disse membraner for i detaljer at observere, hvordan S2- og lipidmembranerne ændrer form, når de interagerer.
Neutroner er også ideelle til denne undersøgelse, fordi de er følsomme over for grundstoffet brint, fælles for alle biologiske molekyler, og dets isotoper. Ved at erstatte nogle brintatomer med deuteriumatomer, videnskabsmænd kan skabe kontrast i deres prøver og selektivt nulstille forskellige strukturelle træk. Denne teknik er nyttig til at studere prøver, der involverer flere komponenter med lignende tætheder, som lipidmembraner.
"Generelt, these membranes are not single-lipid membranes, " said John Ankner, an instrument scientist involved with this study. "They consist of lipids of a certain structure, lipids of another structure, cholesterol, proteiner, and things that come in contact with them."
To capture this complexity, the research team is investigating multiple versions of the membrane, changing the contrast of the sample with deuterium each time.

Researchers at ORNL are using neutron scattering at the Spallation Neutron Source to better understand how spike proteins help the COVID-19 virus infect human cells and what drugs could be effective in stopping them. This research team includes John Ankner (left) and Minh Phan (right). Kredit:ORNL/Genevieve Martin
"By taking multiple measurements and assembling all of this information together, you can create a single picture of how these different components go together, " said Ankner.
The information derived from these experiments will then help steer the team's efforts in selecting and testing drug candidates that could block this interaction, such as fusion inhibitors that successfully blocked original SARS-CoV infections. If these inhibitors can stop the new coronavirus from invading healthy cells, existing drugs could potentially be repurposed to treat COVID-19 patients. The results may also help guide the design of new fusion inhibitors.
Capturing infection
While other studies have used protein crystallography to better understand the atomic structure of the coronavirus S2 subunit alone, this project is analyzing how S2 changes shape when interacting with a lipid membrane. A shape change could be important for inducing actions within a cell after the spike S1 subunit binds to the cell receptor. Phan also notes that the LIQREF instrument allows the team to measure these dynamics under physiological conditions, whereas protein crystallography only allows researchers to capture what the S2 subunit looks like in a crystallized form.
"At ORNL, we have the right tools to study the dynamics of the interaction under physiological conditions. This allows us to better understand how the S2 subunit moves and changes shape naturally in a wet environment, " said Phan. "Such information could complement what experts already know about the protein from crystallography. If we can help verify what this mechanism looks like, then we may have a clearer understanding to guide the development of drugs that block the fusion process.
Collaboration is key
Selvfølgelig, learning more about the S2 subunit and its certain behaviors depends on the ability to grow quality samples, which involves synthesizing S2 subunit proteins, purifying them, and preparing them for experimentation.
Phan and Ankner note that this part of their research has been made possible only through collaboration with labs across ORNL and at outside institutions.
The S2 subunit protein was synthesized in mammalian cell cultures by Steve Foster, a biomedical researcher at the University of Tennessee Medical Center in Knoxville, Tennessee. Through this method, he can develop S2 proteins for research that retain several aspects of its natural structure and function.
"In our lab we routinely use mammalian cell cultures for protein production, so we hope we've produced an S2 protein best suited for this research analysis. Our proximity to ORNL also works well in that the sample doesn't have to travel far, meaning less risk of damaging the protein or distorting its original structure, which is critical for this work, " said Foster.
Following its synthesis, the sample was purified by Jessy Labbé and Michael Melesse Vergara from ORNL's Biosciences Division. Scientists from the ORNL Neutron Sciences Directorate then performed a series of tests to confirm the structure of the sample protein and check its purity. This effort was implemented by Yichong Fan and Wellington Leite from the Bio-Labs team, and Jacob Kinnun and Mary Odom from the SNS team.
"We put an enormous effort into making sure the protein has the right properties going into the experiment. If it does not, we could get spurious results and misinterpret what we're doing, " said Hugh O'Neill, director of ORNL's Center for Structural and Molecular Biology and lead researcher for the Bio-Labs team.
"This virus is extremely delicate in its components, and it's a big challenge to get these materials to the neutron instrument, " said Ankner. "That's why involving various ORNL labs and the University of Tennessee is so crucial. Each step that eventually gets the sample onto our instrument requires the expertise of lots of people."
This project also relied on efforts from the LIQREF instrument staff, who were instrumental in developing the systems, protocols, and modeling frameworks necessary to run the experiments and interpret the data.
"Experts across the division, across ORNL, and from partner institutes have come together for this project, " said Phan. "We couldn't have done this without their support, and it's greater motivation to fulfill our mission."
 Varme artikler
Varme artikler
-
 Nyt værktøj udviklet til at diagnosticere og overvåge autoimmune lidelserKredit:Daria Sokol/MIPT Forskere fra Prokhorov General Physics Institute ved Det Russiske Videnskabsakademi og Moskva Institut for Fysik og Teknologi har udviklet en ny metode til diagnosticering
Nyt værktøj udviklet til at diagnosticere og overvåge autoimmune lidelserKredit:Daria Sokol/MIPT Forskere fra Prokhorov General Physics Institute ved Det Russiske Videnskabsakademi og Moskva Institut for Fysik og Teknologi har udviklet en ny metode til diagnosticering -
 Video:Coronavirus-vaccine:Hvor er vi, og hvad er det næste?Kredit:The American Chemical Society Du har måske hørt, at COVID-19-vaccineforsøg er i gang i Seattle. Hvad er det præcist, der testes? Hvor meget længere vil disse test tage? Og hvornår kan vi f
Video:Coronavirus-vaccine:Hvor er vi, og hvad er det næste?Kredit:The American Chemical Society Du har måske hørt, at COVID-19-vaccineforsøg er i gang i Seattle. Hvad er det præcist, der testes? Hvor meget længere vil disse test tage? Og hvornår kan vi f -
 Syre og base Real-eksemplerSyrer og baser bruges ofte i videnskabslaboratorier i hele landet, men disse magtfulde stoffer har en lang række anvendelser i vores hverdag. Syrer og baser bruges på det industrielle niveau, hvilket
Syre og base Real-eksemplerSyrer og baser bruges ofte i videnskabslaboratorier i hele landet, men disse magtfulde stoffer har en lang række anvendelser i vores hverdag. Syrer og baser bruges på det industrielle niveau, hvilket -
 Modellen lærer, hvordan individuelle aminosyrer bestemmer proteinfunktionenEn ny model udviklet af MIT-forskere skaber rigere, lettere beregnelige repræsentationer af, hvordan individuelle aminosyrer bestemmer et proteins funktion, som kunne bruges til at designe og teste ny
Modellen lærer, hvordan individuelle aminosyrer bestemmer proteinfunktionenEn ny model udviklet af MIT-forskere skaber rigere, lettere beregnelige repræsentationer af, hvordan individuelle aminosyrer bestemmer et proteins funktion, som kunne bruges til at designe og teste ny
- Undersøgelse viser, at flertallet af offentligheden ikke er bekendt med afstemningsforanstaltninger
- Kortvarig tropisk storm Gil giver et spark på NASA-billeder
- Hvordan tror du krig vil være i 2050?
- Ionstråler baner vej for nye slags ventiler til brug i spintronik
- Sådan beregnes vægten af et objekt
- Hvad er augmented reality, alligevel?