Nyt RNA-baseret værktøj kan belyse hjernekredsløb, redigere specifikke celler
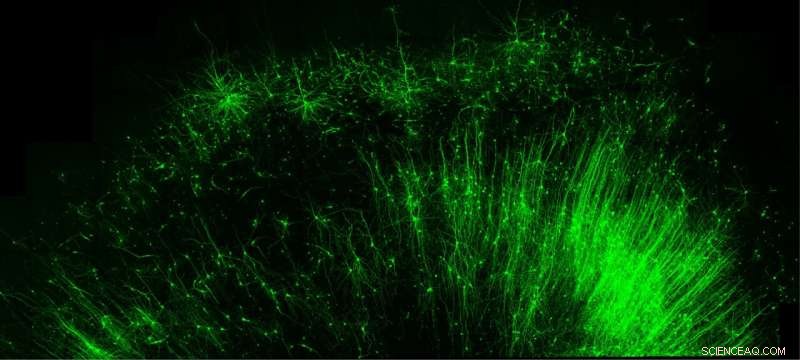
At mærke og belyse kun de hæmmende "bremse"-celler (grønne) i menneskets hjernevæv er blot en af mange ting, det nye værktøj fra Duke University, CellREADR, kan gøre. Kredit:Derek Southwell, Duke University
Duke University-forskere har udviklet et RNA-baseret redigeringsværktøj, der retter sig mod individuelle celler i stedet for gener. Det er i stand til præcist at målrette enhver type celle og selektivt tilføje ethvert protein af interesse.
Forskere sagde, at værktøjet kunne gøre det muligt at modificere meget specifikke celler og cellefunktioner for at håndtere sygdom.
Ved hjælp af en RNA-baseret sonde, et hold ledet af neurobiolog Z. Josh Huang, Ph.D. og postdoc-forsker Yongjun Qian, Ph.D. demonstreret, at de kan indføre fluorescerende tags i celler for at mærke specifikke typer hjernevæv; en lysfølsom tænd/sluk-knap til at dæmpe eller aktivere neuroner efter eget valg; og endda et selvdestruktionsenzym til præcist at fjerne nogle celler, men ikke andre. Værket vises 5. oktober i Nature .
Deres selektive celleovervågnings- og kontrolsystem er afhængige af ADAR-enzymet, som findes i hvert dyrs celler. Selvom disse er tidlige dage for CellREADR (Cell access through RNA sensing by Endogenous ADAR), ser de mulige anvendelser ud til at være uendelige, sagde Huang, ligesom dets potentiale til at arbejde på tværs af dyreriget.
"Vi er begejstrede, fordi dette giver en forenklet, skalerbar og generaliserbar teknologi til at overvåge og manipulere alle celletyper i ethvert dyr," sagde Huang. "Vi kunne faktisk ændre specifikke typer af cellefunktion for at håndtere sygdomme, uanset deres oprindelige genetiske disposition," sagde han. "Det er ikke muligt med nuværende terapier eller medicin."
CellREADR er en tilpasselig streng af RNA, der består af tre hovedsektioner:en sensor, et stopskilt og et sæt tegninger.
Først beslutter forskerholdet, hvilken specifik celletype de ønsker at undersøge, og identificerer et mål-RNA, der er unikt produceret af den celletype. Værktøjets bemærkelsesværdige vævsspecificitet er afhængig af, at hver celletype fremstiller signatur-RNA, der ikke ses i andre celletyper.
En sensorsekvens udformes derefter som mål-RNA's komplementære streng. Ligesom trinene på DNA består af komplementære molekyler, der i sagens natur er tiltrukket af hinanden, har RNA det samme magnetiske potentiale til at forbinde med et andet stykke RNA, hvis det har matchende molekyler.
Efter at en sensor er kommet ind i en celle og har fundet sin mål-RNA-sekvens, glomer begge stykker sammen for at skabe et stykke dobbeltstrenget RNA. Denne nye RNA mashup udløser enzymet ADAR til at inspicere den nye skabelse og derefter ændre et enkelt nukleotid af dets kode.
The ADAR enzyme is a cell-defense mechanism designed to edit double-strand RNA when it occurs, and is believed to be found in all animal cells.
Knowing this, Qian designed CellREADR's stop sign using the same specific nucleotide ADAR edits in double-stranded RNA. The stop sign, which prevents the protein blueprints from being built, is only removed once CellREADR's sensor docks to its target RNA sequence, making it highly specific for a given cell type.
Once ADAR removes the stop sign, the blueprints can be read by cellular machinery that builds the new protein inside the target cell.
In their paper, Huang and his team put CellREADR through its paces. "I remember two years ago when Yongjun built the first iteration of CellREADR and tested it in a mouse brain," Huang said. "To my amazement, it worked spectacularly on his first try."
The team's careful planning and design paid off as they were then able to demonstrate CellREADR accurately labelled specific brain cell populations in living mice, as well as effectively added activity monitors and control switches where directed. It also worked well in rats, and in human brain tissue collected from epilepsy surgeries.
"With CellREADR, we can pick and choose populations to study and really begin to investigate the full range of cell types present in the human brain," said co-author Derek Southwell, M.D., Ph.D., a neurosurgeon and assistant professor in the department of neurosurgery at Duke.
Southwell hopes CellREADR will improve his and others' understanding of the wiring diagram for human brain circuits and the cells within them, and in doing so, help advance new therapies for neurological disorders, such as a promising new method to treat drug-resistant epilepsy he is piloting.
Huang and Qian are especially hopeful about CellREADR's potential as a "programmable RNA medicine" to possibly cure diseases—since that's what drew them both to science in the first place. They have applied for a patent on the technology.
"When I majored in pharmacology as an undergraduate, I was very naïve," Qian said. "I thought you could do a lot of things, like cure cancer, but actually it's very difficult. However, now I think, yes maybe we can do it." + Udforsk yderligere
New technology identifies molecular properties of cells and maps their location within tissues
 Varme artikler
Varme artikler
-
 Er psykiske sygdomme genetiske?Det er ingen hemmelighed, at familier deler gener, men er psykiske sygdomme en del af handlen? © Owen Franken/CORBIS I århundreder, vi har gættet gennem anekdotiske beviser for, at nogle psykiske lid
Er psykiske sygdomme genetiske?Det er ingen hemmelighed, at familier deler gener, men er psykiske sygdomme en del af handlen? © Owen Franken/CORBIS I århundreder, vi har gættet gennem anekdotiske beviser for, at nogle psykiske lid -
 Føderale regering:Ingen truede arter på listen for hvalrosI denne 18. april, 2004, filbillede leveret af U.S. Fish and Wildlife Service, Stillehavshvalrossekøer og -årige hviler på is i Alaska. Trump-administrationen vil ikke tilføje stillehavshvalros til li
Føderale regering:Ingen truede arter på listen for hvalrosI denne 18. april, 2004, filbillede leveret af U.S. Fish and Wildlife Service, Stillehavshvalrossekøer og -årige hviler på is i Alaska. Trump-administrationen vil ikke tilføje stillehavshvalros til li -
 Venstre eller højre? Ligesom mennesker, bier har en præferenceHonningbi (Apis mellifera) lander på en marietidselblomst (Silybum marianum). Kredit:Fir0002/Flagstaffotos/ Wikipedia/GFDL v1.2 En opdagelse af, at bier har individuelle flyveretningspræferencer,
Venstre eller højre? Ligesom mennesker, bier har en præferenceHonningbi (Apis mellifera) lander på en marietidselblomst (Silybum marianum). Kredit:Fir0002/Flagstaffotos/ Wikipedia/GFDL v1.2 En opdagelse af, at bier har individuelle flyveretningspræferencer, -
 Delfiner slår mennesker, chimpanser ved tidlige tegn på selvbevidsthedSpejl, spejl på væggen, hvem er det lyseste pattedyr af dem alle? Brug af spejlbilleder, forskere fandt ud af, at flaskefugle delfiner viser tegn på selvbevidsthed tidligere i livet end mennesker og
Delfiner slår mennesker, chimpanser ved tidlige tegn på selvbevidsthedSpejl, spejl på væggen, hvem er det lyseste pattedyr af dem alle? Brug af spejlbilleder, forskere fandt ud af, at flaskefugle delfiner viser tegn på selvbevidsthed tidligere i livet end mennesker og
- Forskere finder, at stress under graviditeten påvirker barnets størrelse
- Kan et Wi-Fi-netværk nogensinde være helt sikkert?
- Seks klimaforandringsløsninger kan vi alle blive enige om
- Sådan beregnes en ændring i Momentum
- Jeg bygger matematiske programmer, der kan opdage fremtidens stoffer
- Flere beviser for vand på Mars